Statistics for Soil Survey - Part 1
2025-05-01
Pre-course Assignment

0.1 Create Workspace
Make a local folder C:\workspace2 to use as a working directory for this course. Use all lower case letters please.
0.2 Configure RStudio
Open RStudio, and edit the “Global Options” (Main menu: Tools → Global Options).
0.3 Essentials
These options are important for pleasant, reproducible and efficient use of the RStudio environment:
Change the default working directory to
C:\workspace2(R General Tab)Uncheck “Restore .Rdata into workspace at startup” (R General Tab) VERY IMPORTANT
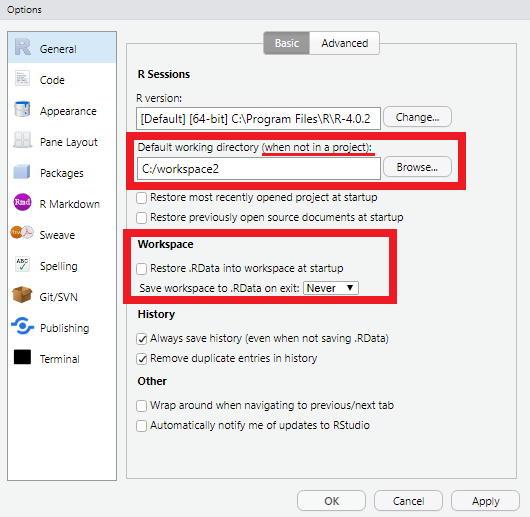
RStudio detects the available R installations on your computer.
Individual versions are certified for the Software Center as they become available, and sometimes there is a more recent version available for download. It is worth taking the time before installing packages to get the latest version of R available to you. This is to minimize compatibility issues which arise over time.
0.4 Personalization
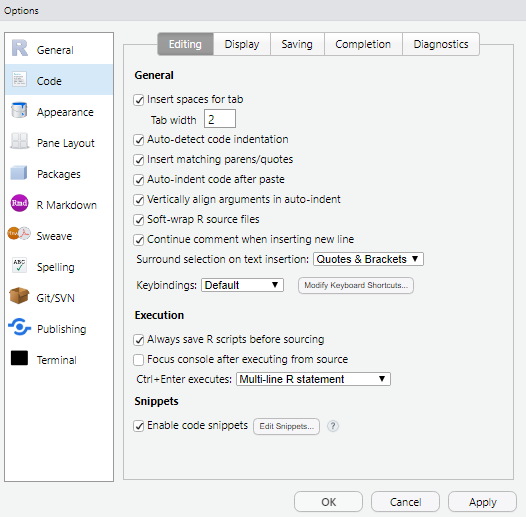
Optional: Check “Soft-wrap R source files” (Code/Editing Tab)
Optional: Show help tooltips, control auto-completion and diagnostics (Code/Completion and Diagnostics Tabs)
Optional: Update code font size, colors and theme (Appearance)
Optional: Use RStudio Projects (top-right corner) to manage working directories
0.5 Install Required Packages
Packages can be installed by name from the Comprehensive R Archive Network (CRAN) using the base R function install.packages(). There are a lot of packages out there–many more than you will download here, and many of which are useful for Soil Survey work.
The first time you install packages, R may ask you if you want to create a local package library. You need to do this because we cannot write to system folders as non-administrator users on CCE machines. The default location for R package library on Windows is: C:\Users\<User.Name>\AppData\Local\R\win-library\<X.X> where <User.Name> is the current Windows user name and <X.X> is the version of R packages are being installed for.
If you have an existing R package library (for same minor version of R), you can copy that library into the AppData\Local\R folder as needed.
For example, to download and install the remotes package from CRAN:
To install the R packages used in this class copy all of the code from the box below and paste into the R console window. Paste after the command prompt (>) and press enter.
Downloading and configuring the packages will take a while if you are installing or upgrading all of the packages in the list below.
On particularly slow network connections, i.e. over VPN or USDA network in general, it may be necessary to increase the “timeout” option to ensure the downloads have sufficient time to complete.
# increase default timeout from 1 minute to 5 minutes (for current session only)
options(timeout = 300)## character vector of package names
packages <- c(
# soil
"aqp", "soilDB", "sharpshootR", "soiltexture",
# gis
"raster", "sp", "sf", "terra", "gdalUtilities",
"rgrass", "RSAGA", "exactextractr", "fasterize",
# data management
"dplyr", "tidyr", "devtools", "roxygen2", "Hmisc", "circular", "DT", "remotes", "DescTools",
# databases
"DBI", "odbc", "RSQLite",
# graphics
"ggplot2", "latticeExtra", "maps", "spData", "tmap", "kableExtra", "corrplot", "farver",
"mapview", "ggmap", "plotrix", "rpart.plot", "visreg", "diagram", "GGally", "wesanderson",
"viridisLite", "prettymapr",
# modeling
"car", "rms", "randomForest", "ranger", "party",
"caret", "vegan", "ape", "shape", "modEvA", "gower", "MBESS", "yardstick",
# sampling
"clhs", "spcosa", "sgsR"
)
# ipkCRAN: a helper fuction for installing required packages from CRAN
source("https://raw.githubusercontent.com/ncss-tech/stats_for_soil_survey/master/ipkCRAN.R")
## - p: vector of package names
## - up: logical - upgrade installed packages? Default: TRUE
## up = TRUE to download all packages
## up = FALSE to download only packages you don't already have installed
ipkCRAN(p = packages, up = TRUE)The ipkCRAN function will let you know if any of the above packages fail to install.
Whenever you run some code always check the console output for warnings and errors before continuing. It may be easiest to send commands individually to learn about and inspect their output, rather than running the entire file and wondering where an error occurred.
0.6 Dealing With Errors
If a lot of output is produced by a command you should scroll up and sift through it as best you can. Copy and paste parts of the error message to use in internet searches, and try to find cases where folks have encountered problems.
0.6.1 No output is produced after pasting into console
If you do not have a new command prompt (>) and a blinking cursor on the left hand side of your console, but instead see a + after you run a command, R may think you are still in the middle of submitting input to the “read-eval-print-loop” (REPL).
If this is not expected you are possibly missing closing quotes, braces, brackets or parentheses. R needs to know you were done with your expression, so you may need to supply some input to get the command to be complete.
Pasting code line-by-line is useful but prone to input errors with multiline expressions. Alternately, you can run commands or an entire file using the GUI or keyboard shortcuts such as Ctrl+Enter. You have a chance to try this in the example at the end.
0.6.2 ‘SOMEPACKAGE’ is not available (for R version X.Y.Z)
This means either:
A package named ‘SOMEPACKAGE’ exists but it is not available for your version of R
CRAN does not have a package with that name
You can try again, but first check for spelling and case-sensitivity. When in doubt search the package name on Google or CRAN to make sure you have it right.
Note that not all R packages are available on CRAN: there are many other ways that you can deliver packages (including GitHub described below).
0.7 Packages not on CRAN
Some R packages rely on compiled code. Windows users are limited to installing “binary” versions of such packages from CRAN unless they have “Rtools” installed. The Rtools software is available from the Software Center, and is specific to the version of R you have.
One way to get the latest binary builds of R packages that use compiled code is by using https://r-universe.dev. This website provides custom repositories that can be used in addition to the defaults in install.packages()
For example, you can install raster-related “rspatial” packages from r-universe.dev. This may not be “required” but it is good to know how to specify an alternate package repository source using the repos= argument. Check with your mentor to see if there are known issues with current CRAN packages.
To install the latest version of packages from the Algorithms for Quantitative Pedology (AQP) suite off GitHub we use the remotes package.
The AQP packages are updated much more frequently on GitHub than they are on CRAN.
Generally, the CRAN versions (installed above) are the “stable” releases whereas the GitHub repositories have new features and bug fixes.
remotes::install_github("ncss-tech/aqp", dependencies=FALSE, upgrade=FALSE, build=FALSE)
remotes::install_github("ncss-tech/soilDB", dependencies=FALSE, upgrade=FALSE, build=FALSE)
remotes::install_github("ncss-tech/sharpshootR", dependencies=FALSE, upgrade=FALSE, build=FALSE)
remotes::install_github("ncss-tech/soilReports", dependencies=FALSE, upgrade=FALSE, build=FALSE)0.8 Install NASIS
If you do not have NASIS installed you will not be able to complete the next steps. For USDA staff NASIS can be installed from the Software Center. Search "NASIS" and you should find two applications. First you must install “Microsoft SQL Server Express for the NASIS Client”, then restart your computer, and then install the “NASIS Client”.
If you do not have access to the Software Center, or cannot find NASIS, it can be installed by OCIO staff by submitting an IT support ticket for “Install / Uninstall Software”. If you are not a USDA employee you can find additional information on NASIS installation here: https://new.cloudvault.usda.gov/index.php/s/xFTJabHiT45WDom
More information on installing and initializing your NASIS client for the first time can be found in the training materials for NASIS 1010 (Basic NASIS) course: https://ncss-tech.github.io/nasis_training/content/pre-course.html
0.9 Connect Local NASIS
Establish an ODBC connection to NASIS by following the directions at the following hyperlink (ODBC Connection to NASIS).
Once you’ve successfully established a ODBC connection, prove it by loading your NASIS selected set with the site and pedon tables for any pedons from your local area. You only need a few pedons at a minimum for this demo – too many (say, >20) will make the example profile plot cluttered.
Paste the below code at the command prompt (
>) and press enter, as you did above.Or create a new R script (Main menu: File → New File → R Script) and paste code into the “Source” pane (script editor window). Then, click the Run button in the top-right corner of the Script Editor or use Ctrl+Enter to run code at the cursor location / any selected code. This will execute the code in the Console.
Submit the resulting plot to your mentor (from “Plot” pane (bottom-right): Export → Save as PDF…)
# load packages into the current session
library(aqp) # provides "SoilProfileCollection" object & more
library(soilDB) # provides database access methods
# get pedons from NASIS selected set
test <- fetchNASIS(from = 'pedons')
# inspect the result
str(test, max.level = 2)
# make a profile plot
# set margins smaller than default
par(mar=c(1,1,1,1))
# make profile plot of selected set, with userpedonid as label
plot(test, label='upedonid')0.10 Proof
Follow the one line example below, copy the output, and submit the results to your mentor. This will help us to verify that all of the required packages have been installed.