Create an adjacency matrix from a data.frame of component data
Source:R/component.adj.matrix.R
component.adj.matrix.RdCreate an adjacency matrix from SSURGO component data
component.adj.matrix(
d,
mu = "mukey",
co = "compname",
wt = "comppct_r",
method = c("community.matrix", "occurrence"),
standardization = "max",
metric = "jaccard",
rm.orphans = TRUE,
similarity = TRUE,
return.comm.matrix = FALSE
)Arguments
- d
data.frame, typically of SSURGO data- mu
name of the column containing the map unit ID (typically 'mukey')
- co
name of the column containing the component ID (typically 'compname')
- wt
name of the column containing the component weight percent (typically 'comppct_r')
- method
one of either:
community.matrix, oroccurrence; see details- standardization
community matrix standardization method, passed to
vegan::decostand- metric
community matrix dissimilarity metric, passed to
vegan::vegdist- rm.orphans
logical, should map units with a single component be omitted? (typically yes)- similarity
logical, return a similarity matrix? (if
FALSE, a distance matrix is returned)- return.comm.matrix
logical, return pseudo-community matrix? (if
TRUEno adjacency matrix is created)
Value
A similarity or adjacency matrix suitable for use with igraph functions or anything else that can accommodate a similarity matrix.
Examples
# \donttest{
if (requireNamespace("igraph") && requireNamespace("vegan")) {
# load sample data set
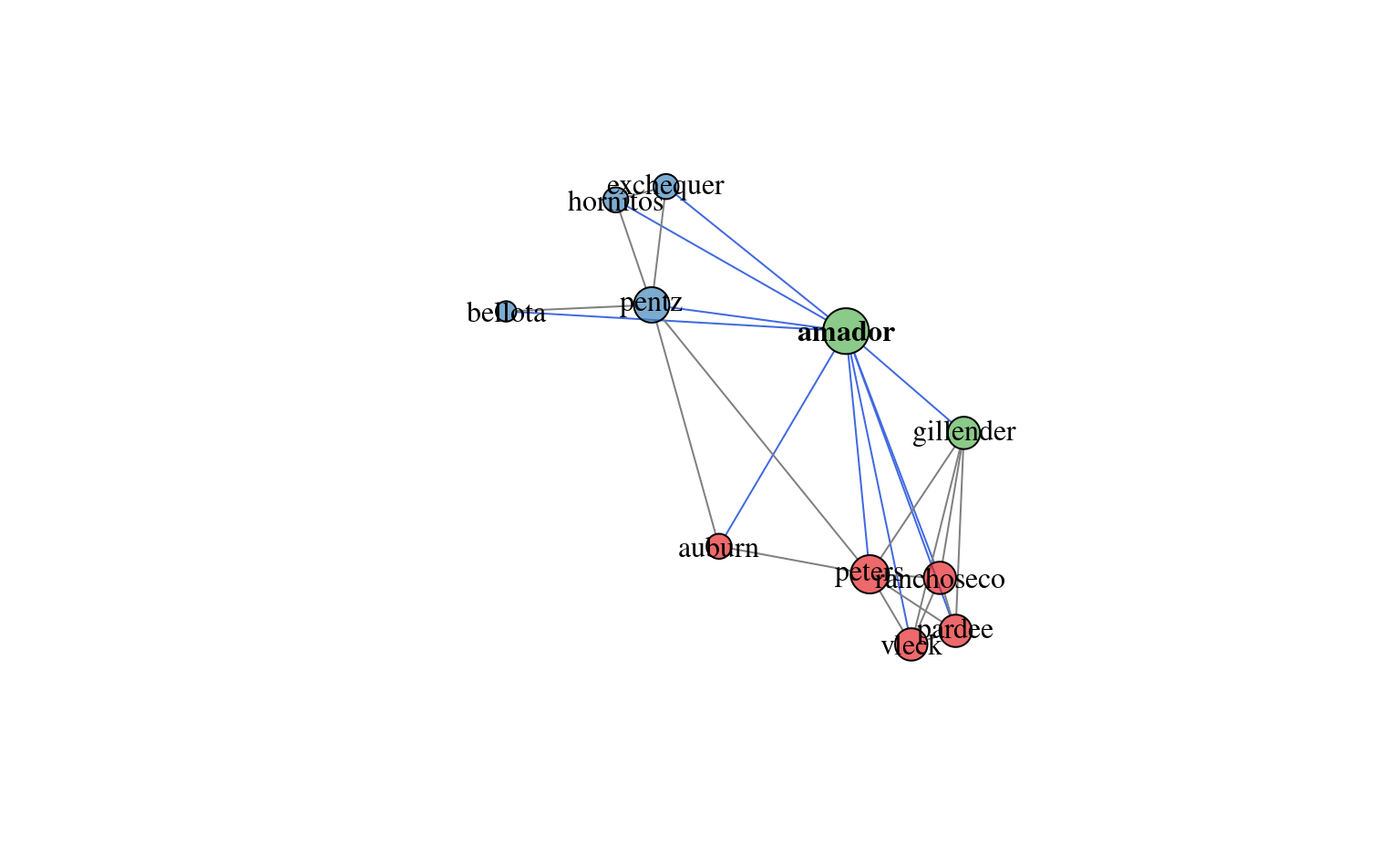
data(amador)
# convert into adjacency matrix
m <- component.adj.matrix(amador)
# plot network diagram, with Amador soil highlighted
plotSoilRelationGraph(m, s = 'amador')
}
#> Loading required namespace: igraph
#> Loading required namespace: vegan
 # }
# }