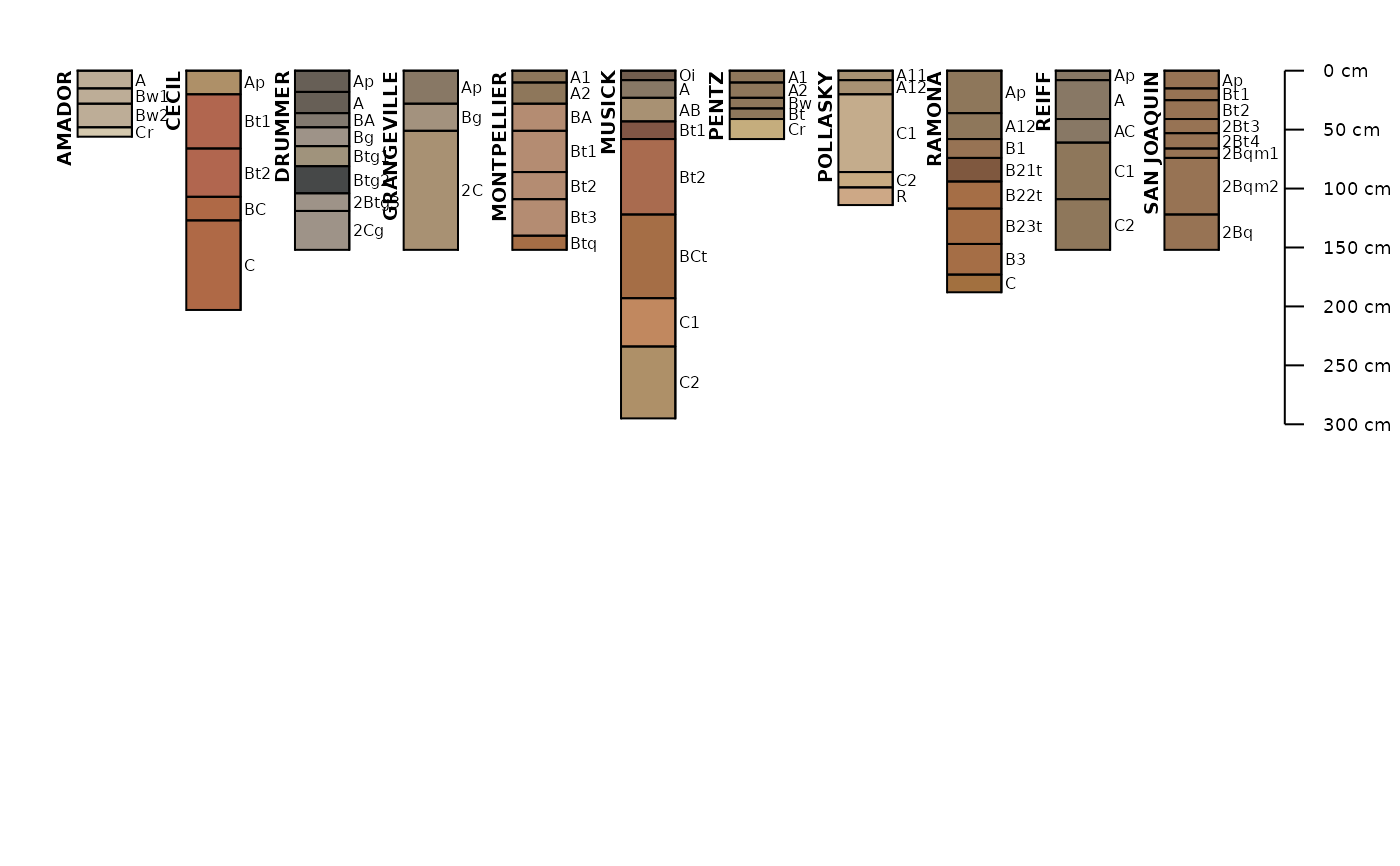
This function fetches a variety of data associated with named soil series, extracted from the USDA-NRCS Official Series Description text files and detailed soil survey (SSURGO). These data are updated quarterly and made available via SoilWeb. Set extended = TRUE and see the soilweb.metadata list element for information on when the source data were last updated.
Value
a SoilProfileCollection object containing basic soil morphology and taxonomic information, a list when extended = TRUE.
Details
The standard set of "site" and "horizon" data are returned as aSoilProfileCollection object (extended = FALSE). The "extended" suite of summary data can be requested by setting extended = TRUE. The resulting object will be a list with the following elements:- SPC
SoilProfileCollectioncontaining standards "site" and "horizon" data- competing
competing soil series from the SC database snapshot
- geog_assoc_soils
geographically associated soils, extracted from named section in the OSD
- geomcomp
empirical probabilities for geomorphic component, derived from the current SSURGO snapshot
- hillpos
empirical probabilities for hillslope position, derived from the current SSURGO snapshot
- mtnpos
empirical probabilities for mountain slope position, derived from the current SSURGO snapshot
- terrace
empirical probabilities for river terrace position, derived from the current SSURGO snapshot
- flats
empirical probabilities for flat landscapes, derived from the current SSURGO snapshot
- shape_across
empirical probabilities for surface shape (across-slope) from the current SSURGO snapshot
- shape_down
empirical probabilities for surface shape (down-slope) from the current SSURGO snapshot
- pmkind
empirical probabilities for parent material kind, derived from the current SSURGO snapshot
- pmorigin
empirical probabilities for parent material origin, derived from the current SSURGO snapshot
- geomorphons
geomorphons landform classification (CONUS only), derived from the current gSSURGO snapshot and a 30m CONUS geomoprhons grid, details pending
- mlra
empirical MLRA membership values, derived from the current SSURGO snapshot
- ecoclassid
area cross-tabulation of ecoclassid by soil series name, derived from the current SSURGO snapshot, major components only
- climate
climate summaries from PRISM stack (CONUS only)
- NCCPI
select quantiles of NCCPI and Irrigated NCCPI, derived from the current SSURGO snapshot
- metadata
metadata associated with SoilWeb cached summaries
extended = TRUE, there are a couple of scenarios in which series morphology contained in SPC do not fully match records in the associated series summary tables (e.g. competing).- 1. A query for soil series that exist entirely outside of CONUS (e.g. PALAU).
- Climate summaries are empty
data.framebecause these summaries are currently generated from PRISM. We are working on a solution that uses DAYMET.- 2. A query for data within CONUS, but OSD morphology missing due to parsing error (e.g. formatting, typos).
- Extended summaries are present but morphology missing from
SPC. A warning is issued.
Note
Requests to the SoilWeb API are split into batches of 100 series names from soils via makeChunks().
References
USDA-NRCS OSD search tools: https://soilseries.sc.egov.usda.gov/
Jasiewicz, J., Stepinski, T., 2013, Geomorphons - a pattern recognition approach to classification and mapping of landforms, Geomorphology, vol. 182, 147-156. (doi:10.1016/j.geomorph.2012.11.005 )
Examples
# \donttest{
library(aqp)
# soils of interest
s.list <- c('musick', 'cecil', 'drummer', 'amador', 'pentz',
'reiff', 'san joaquin', 'montpellier', 'grangeville', 'pollasky', 'ramona')
# fetch and convert data into an SPC
s.moist <- fetchOSD(s.list, colorState='moist')
s.dry <- fetchOSD(s.list, colorState='dry')
# plot profiles
# moist soil colors
par(mar=c(0,0,0,0), mfrow=c(2,1))
aqp::plotSPC(
s.moist,
name = 'hzname',
cex.names = 0.85,
depth.axis = list(line = -4)
)
aqp::plotSPC(
s.dry,
name = 'hzname',
cex.names = 0.85,
depth.axis = list(line = -4)
)
 # extended mode: return a list with SPC + summary tables
x <- fetchOSD(s.list, extended = TRUE, colorState = 'dry')
par(mar=c(0,0,1,1))
if (!is.null(x$SPC) && inherits(x$SPC, "SoilProfileCollection")){
aqp::plotSPC(x$SPC)
}
str(x, 1)
#> List of 19
#> $ SPC :Formal class 'SoilProfileCollection' [package "aqp"] with 8 slots
#> $ competing :'data.frame': 84 obs. of 3 variables:
#> $ geog_assoc_soils:'data.frame': 80 obs. of 2 variables:
#> $ geomcomp :'data.frame': 11 obs. of 9 variables:
#> $ hillpos :'data.frame': 11 obs. of 8 variables:
#> $ mtnpos :'data.frame': 1 obs. of 9 variables:
#> $ terrace :'data.frame': 9 obs. of 5 variables:
#> $ flats :'data.frame': 5 obs. of 7 variables:
#> $ shape_across :'data.frame': 11 obs. of 8 variables:
#> $ shape_down :'data.frame': 11 obs. of 8 variables:
#> $ pmkind :'data.frame': 18 obs. of 5 variables:
#> $ pmorigin :'data.frame': 32 obs. of 5 variables:
#> $ geomorphons :'data.frame': 11 obs. of 12 variables:
#> $ mlra :'data.frame': 47 obs. of 4 variables:
#> $ ecoclassid :'data.frame': 59 obs. of 5 variables:
#> $ climate.annual :'data.frame': 88 obs. of 12 variables:
#> $ climate.monthly :'data.frame': 264 obs. of 14 variables:
#> $ NCCPI :'data.frame': 11 obs. of 16 variables:
#> $ soilweb.metadata:'data.frame': 28 obs. of 2 variables:
# }
# extended mode: return a list with SPC + summary tables
x <- fetchOSD(s.list, extended = TRUE, colorState = 'dry')
par(mar=c(0,0,1,1))
if (!is.null(x$SPC) && inherits(x$SPC, "SoilProfileCollection")){
aqp::plotSPC(x$SPC)
}
str(x, 1)
#> List of 19
#> $ SPC :Formal class 'SoilProfileCollection' [package "aqp"] with 8 slots
#> $ competing :'data.frame': 84 obs. of 3 variables:
#> $ geog_assoc_soils:'data.frame': 80 obs. of 2 variables:
#> $ geomcomp :'data.frame': 11 obs. of 9 variables:
#> $ hillpos :'data.frame': 11 obs. of 8 variables:
#> $ mtnpos :'data.frame': 1 obs. of 9 variables:
#> $ terrace :'data.frame': 9 obs. of 5 variables:
#> $ flats :'data.frame': 5 obs. of 7 variables:
#> $ shape_across :'data.frame': 11 obs. of 8 variables:
#> $ shape_down :'data.frame': 11 obs. of 8 variables:
#> $ pmkind :'data.frame': 18 obs. of 5 variables:
#> $ pmorigin :'data.frame': 32 obs. of 5 variables:
#> $ geomorphons :'data.frame': 11 obs. of 12 variables:
#> $ mlra :'data.frame': 47 obs. of 4 variables:
#> $ ecoclassid :'data.frame': 59 obs. of 5 variables:
#> $ climate.annual :'data.frame': 88 obs. of 12 variables:
#> $ climate.monthly :'data.frame': 264 obs. of 14 variables:
#> $ NCCPI :'data.frame': 11 obs. of 16 variables:
#> $ soilweb.metadata:'data.frame': 28 obs. of 2 variables:
# }