This function generates Newick tree format strings for a single tree. Taxa are assigned relative positions within their parent to indicate the order that they "key out."
Arguments
- x
Optional: a taxon name to get children of.
- level
Level to build the tree at. One of
"suborder","greatgroup","subgroup". Defaults to"suborder"whenxis not specified. Whenxis specified butlevelis not specified,levelis calculated fromtaxon_to_level(x).- what
Either
"taxon"(default; for taxon names (quoted for subgroups)) or"code"
Details
The output from this function is a character string with parenthetical format encoding a single tree suitable for input into functions such as ape::read.tree(). Multiple trees can be combined together in the file or text string supplied to your tree-parsing function of choice.
Examples
if (requireNamespace("ape")) {
par(mar = c(0, 0, 0, 0))
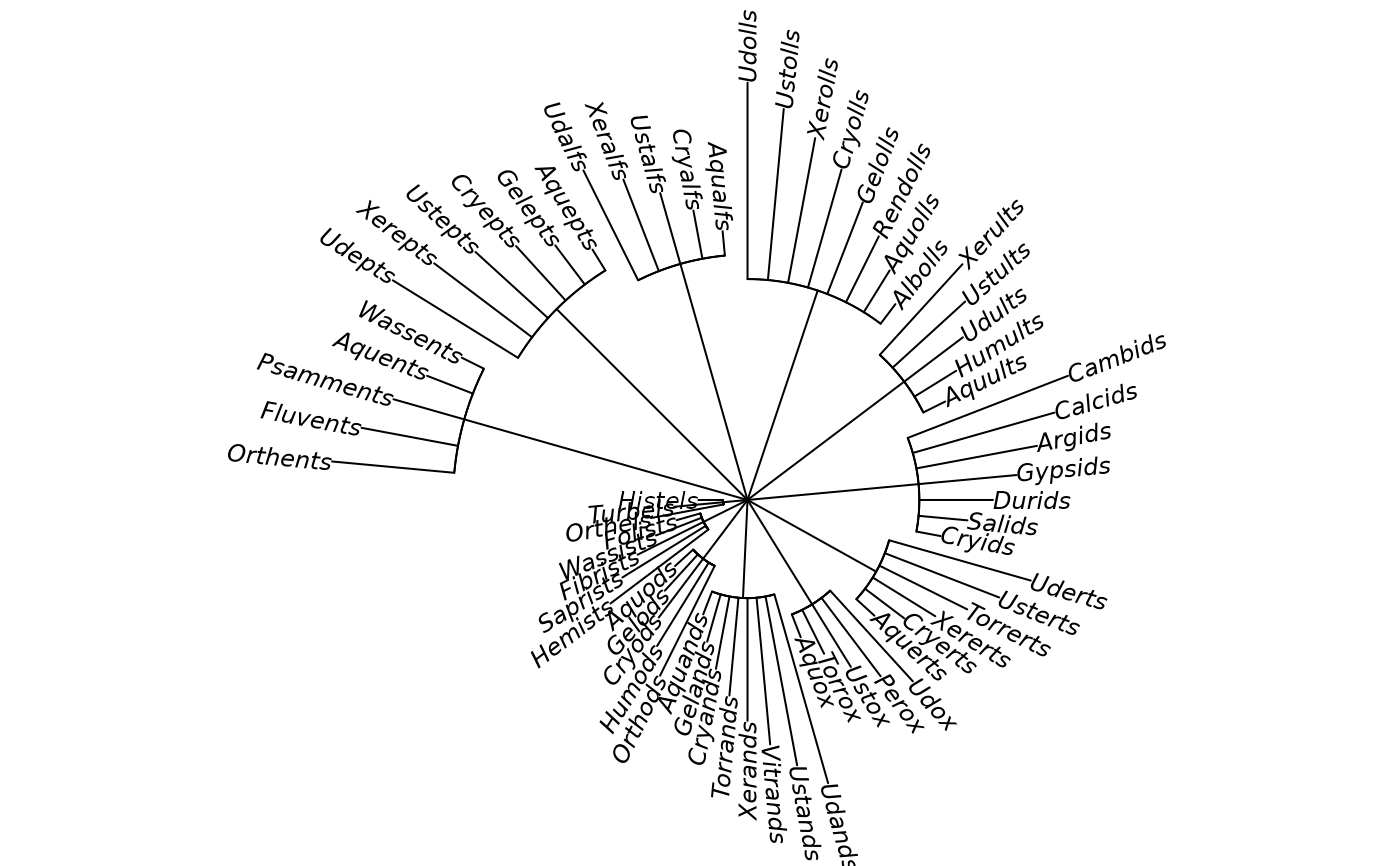
# "fan"
mytr <- ape::read.tree(text = newick_string(level = "suborder"))
plot(mytr, "f", rotate.tree = 180, cex = 0.75)
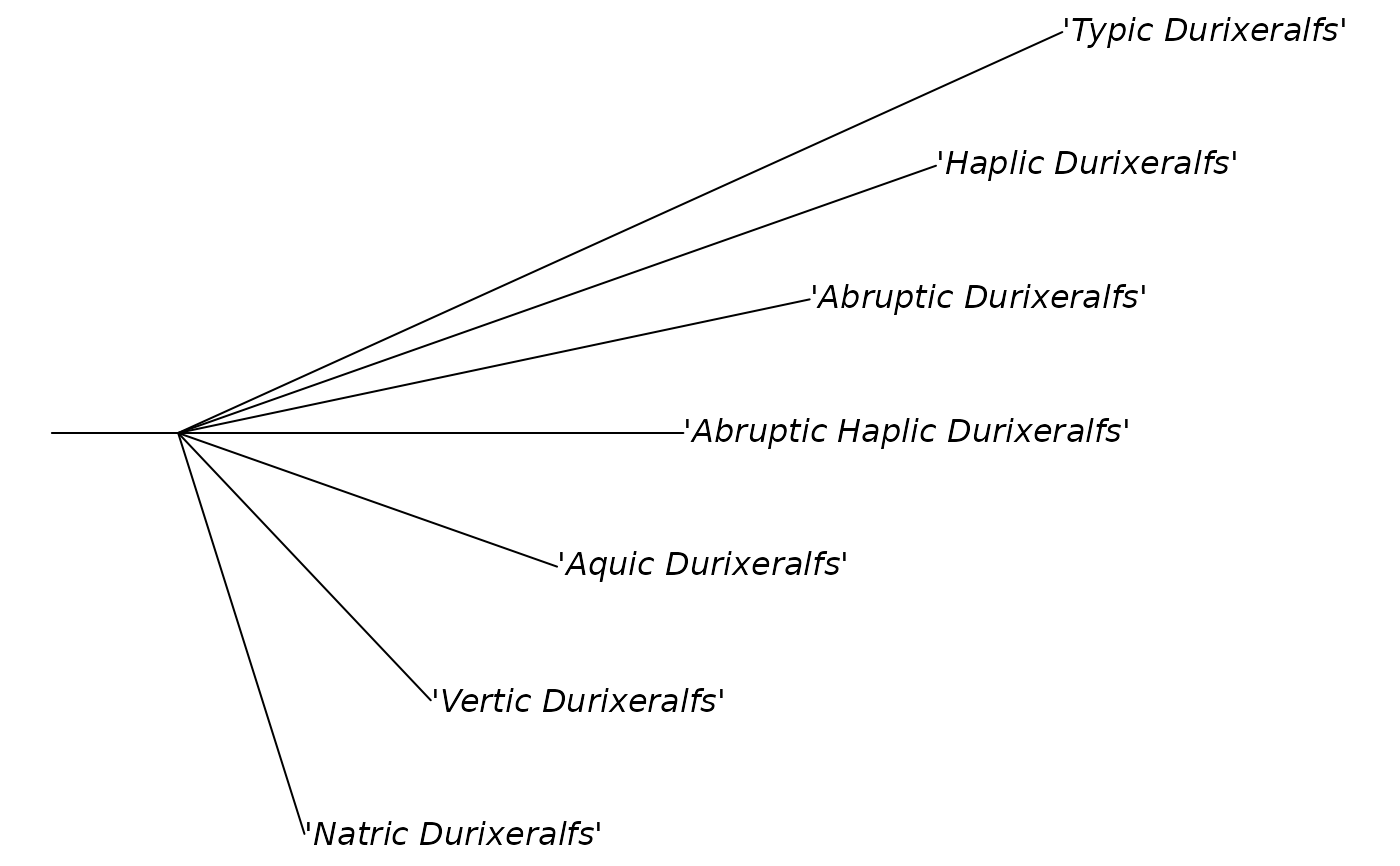
# "cladogram"
mytr <- ape::read.tree(text = newick_string("durixeralfs", level = "subgroup"))
plot(mytr, "c")
# "cladogram" (using taxon codes instead of subgroups)
mytr <- ape::read.tree(text = newick_string("xeralfs", level = "subgroup", what = "code"))
plot(mytr, "c")
dev.off()
}
#> Loading required namespace: ape
 #> null device
#> 1
#> null device
#> 1