Run a Rule on Custom Property Data
interpret.RdAllows for evaluation of a primary rule including subrules, operators, hedges, and evaluations.
Usage
interpret(x, propdata, ...)
# S4 method for class 'Node,data.frame'
interpret(x, propdata, mode = "node", cache = FALSE, ...)
# S4 method for class 'character,data.frame'
interpret(x, propdata, mode = "node", cache = FALSE, ...)
# S4 method for class 'Node,SpatRaster'
interpret(
x,
propdata,
mode = "node",
cores = 1,
core_thresh = 250000L,
file = paste0(tempfile(), ".tif"),
nrows = nrow(propdata)/(terra::ncell(propdata)/core_thresh),
overwrite = TRUE,
...
)
# S4 method for class 'character,SpatRaster'
interpret(
x,
propdata,
mode = "node",
cores = 1,
core_thresh = 250000L,
file = paste0(tempfile(), ".tif"),
nrows = nrow(propdata)/(terra::ncell(propdata)/core_thresh),
overwrite = TRUE,
...
)Arguments
- x
A data.tree object containing rule tree, or a character giving the rule name to load with
initRuleset().- propdata
data.frame or SpatRaster object with column names corresponding to input properties. The column names should be named using
make.names(propname)wherepropnameis the property name from NASIS_properties data object.- ...
Additional arguments
- mode
character. Either
"table"or"node"(default) . Controls back-end rating calculation method using base R or data.tree, respectively.- cache
logical. Save input property data in data.tree object? Default:
FALSE- cores
integer. Default
1core.- core_thresh
integer. Default
250000cells.- file
character. Path to output raster file. Defaults to a temporary GeoTIFF.
- nrows
integer. Default
nrow(propdata) / (terra::ncell(propdata) / core_thresh)- overwrite
logical. Overwrite
fileif it exists?
Value
data.frame containing "rating". When mode="node" the input object x is modified in place as a side effect with "rating" values. When cache=TRUE the input "data" values are also stored within each node.
Examples
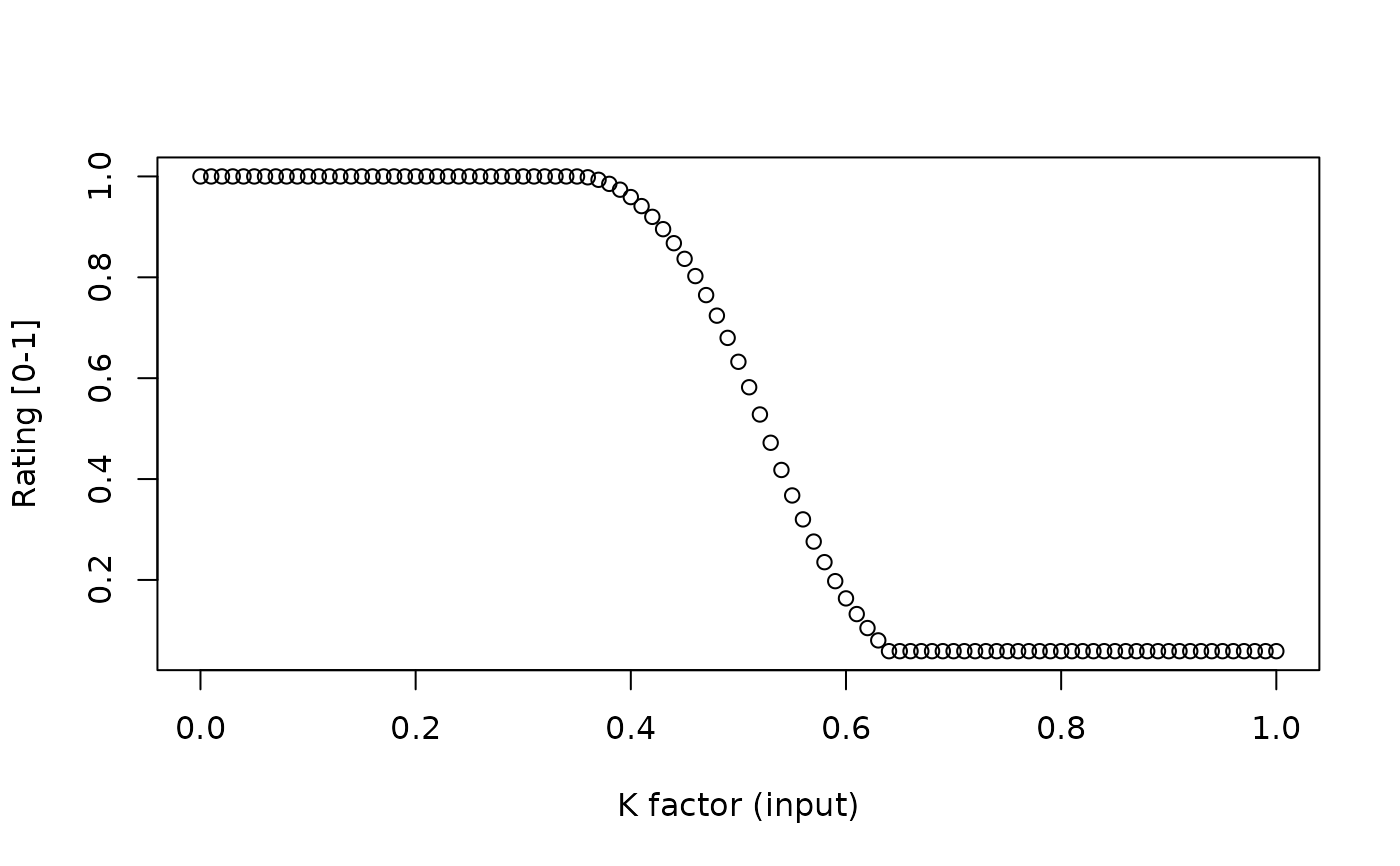
r <- initRuleset("Erodibility Factor Maximum")
p <- getPropertySet(r)
my_data <- data.frame(Kmax = seq(0, 1, 0.01))
colnames(my_data) <- make.names(p$propname)
res <- interpret(r, my_data)
plot(res$rating ~ my_data[[1]],
xlab = "K factor (input)",
ylab = "Rating [0-1]")